 Lumba S, Tsuchiya Y, Delmas F, Hezky J, Provart NJ, Shi Lu Q, McCourt P, Gazzarrini S
Lumba S, Tsuchiya Y, Delmas F, Hezky J, Provart NJ, Shi Lu Q, McCourt P, Gazzarrini S
BMC Biol. 2012 ;10():8
PubMed PMID: 22348746
Abstract
BACKGROUND:
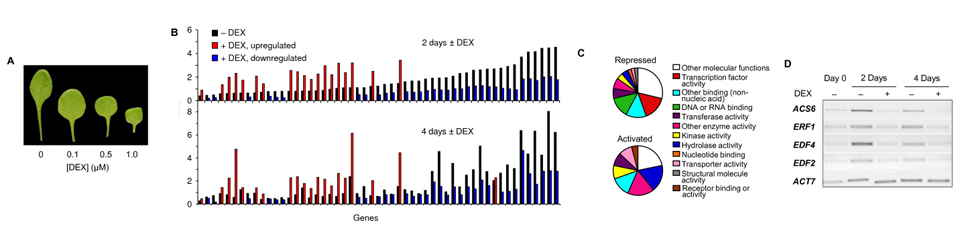
The embryonic temporal regulator FUSCA3 (FUS3) plays major roles in the establishment of embryonic leaf identity and the regulation of developmental timing. Loss-of-function mutations of this B3 domain transcription factor result in replacement of cotyledons with leaves and precocious germination, whereas constitutive misexpression causes the conversion of leaves into cotyledon-like organs and delays vegetative and reproductive phase transitions.
RESULTS:
Herein we show that activation of FUS3 after germination dampens the expression of genes involved in the biosynthesis and response to the plant hormone ethylene, whereas a loss-of-function fus3 mutant shows many phenotypes consistent with increased ethylene signaling. This FUS3-dependent regulation of ethylene signaling also impinges on timing functions outside embryogenesis. Loss of FUS3 function results in accelerated vegetative phase change, and this is again partially dependent on functional ethylene signaling. This alteration in vegetative phase transition is dependent on both embryonic and vegetative FUS3 function, suggesting that this important transcriptional regulator controls both embryonic and vegetative developmental timing.
CONCLUSION:
The results of this study indicate that the embryonic regulator FUS3 not only controls the embryonic-to-vegetative phase transition through hormonal (ABA/GA) regulation but also functions postembryonically to delay vegetative phase transitions by negatively modulating ethylene-regulated gene expression.

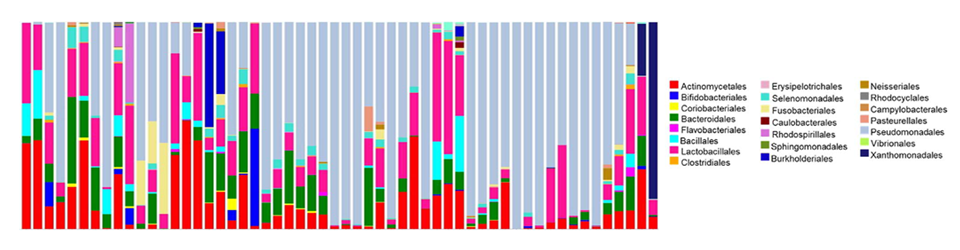 Maughan H, Cunningham KS, Wang PW, Zhang Y, Cypel M, Chaparro C, Tullis DE, Waddell TK, Keshavjee S, Liu M, Guttman DS, Hwang DM
Maughan H, Cunningham KS, Wang PW, Zhang Y, Cypel M, Chaparro C, Tullis DE, Waddell TK, Keshavjee S, Liu M, Guttman DS, Hwang DM Patel RV, Nahal HK, Breit R, Provart NJ
Patel RV, Nahal HK, Breit R, Provart NJ Austin RS, Vidaurre D, Stamatiou G, Breit R, Provart NJ, Bonetta D, Zhang J, Fung P, Gong Y, Wang PW, McCourt P, Guttman DS
Austin RS, Vidaurre D, Stamatiou G, Breit R, Provart NJ, Bonetta D, Zhang J, Fung P, Gong Y, Wang PW, McCourt P, Guttman DS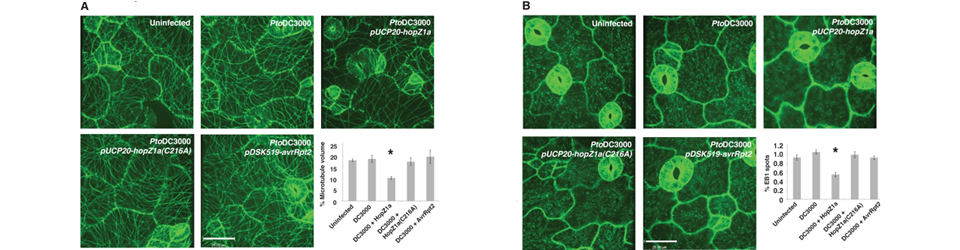 Lee AH, Hurley B, Felsensteiner C, Yea C, Ckurshumova W, Bartetzko V, Wang PW, Quach V, Lewis JD, Liu YC, Börnke F, Angers S, Wilde A, Guttman DS, Desveaux D
Lee AH, Hurley B, Felsensteiner C, Yea C, Ckurshumova W, Bartetzko V, Wang PW, Quach V, Lewis JD, Liu YC, Börnke F, Angers S, Wilde A, Guttman DS, Desveaux D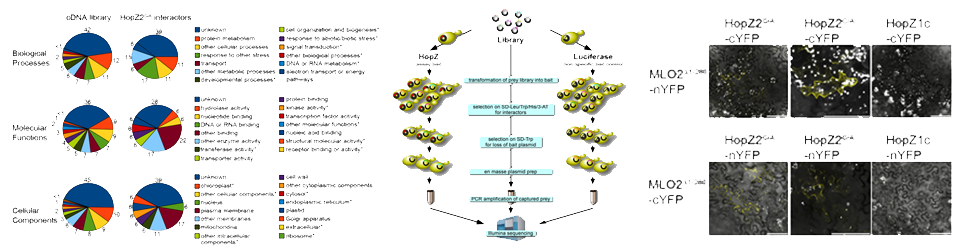 Lewis JD, Wan J, Ford R, Gong Y, Fung P, Nahal H, Wang PW, Desveaux D, Guttman DS
Lewis JD, Wan J, Ford R, Gong Y, Fung P, Nahal H, Wang PW, Desveaux D, Guttman DS